Medical Biochemistry
General Facts
Research
Selected Publications
Selected Funding, Collaboration
Keywords: Cell Cycle Control, Cell Proliferation; Protein Kinases, Translational Control, Toxicology, Immunometabolism, Protein Microanalysis Facility, Mass Spectrometry, capillary electrophoresis
Research (ÖSTAT Classification) : 104002, 106002, 106037, 106052, 3013, 301303, 301904, 301211, 301114
General Facts
The institute currently hosts two research groups led by Ludger Hengst and Johanna Gostner and a protein core facility led by Bettina Sarg. Using a broad range of cell biological and molecular biological techniques, the cell cycle and cell proliferation group investigates molecular mechanisms that govern the decision between cell proliferation and cell cycle exit to quiescence or differentiation in normal mammalian cells and tissues and in cancer models. Here, we focus on the regulation of cyclin-dependent kinase (CDK) activities and CDK inhibitors and we study mechanisms of transcriptional, translational and post-translational control. The biochemical immunotoxicology group focuses on cellular responses to chemical exposure and its immunometabolic consequences. The protein core facility develops and establishes novel analytical methods and technologies. This facility provides a wide platform of services such as HPLC, capillary electrophoresis and MS-based analysis and quantification of proteins from cells, plasma or tissues, as well as elemental analysis using graphite furnace atomic absorption spectrometry (GF-AAS) to measure specific elements in tissue biopsies, blood or urine.
Research
Cell Cycle and Cell Proliferation
Ludger Hengst
Precisely coordinated cell division and differentiation processes are essential for the growth, development and integrity of multicellular organisms. Before cells commit to division or quiescence, they are exposed to a flood of diverse and sometimes conflicting signals that aim to regulate cell growth, differentiation, cell proliferation or cell fate. Multiple external as well as internal signals can impinge on the central cell cycle control machinery, in order to promote or block cell proliferation. These signals all need to be properly processed and integrated, in order to control cell proliferation and quiescence and to maintain body and organ homeostasis. Incorrect signal interpretation, processing or integration can lead to hypo- or hyperproliferative disorders, including diseases such as cancer or anaemia.
The decision to continue proliferation or to exit from the cell cycle into quiescence is usually made during a specific window of the eukaryotic cell division cycle. This cell cycle can be subdivided into four phases: gap phases G1 and G2 separate DNA replication during the S-phase from segregation of the duplicated DNA and other cellular components in the M-phase (mitosis and cytokinesis). Cells can decide to withdraw from proliferation or to commit to another round of cell division until they progress over the so-called restriction point, a specific point in phase G1 (Fig. 1). Progression over the restriction point renders cells mitogen-independent and they become committed to undergoing another complete cell cycle including S-phase and mitosis.
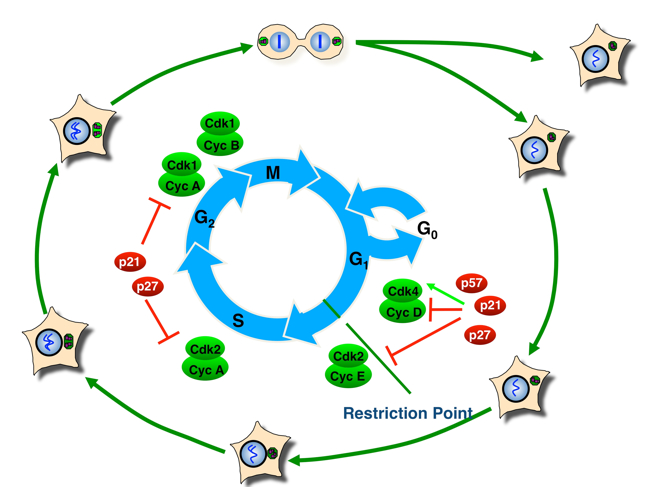
Fig. 1: Overview of the mammalian cell cycle. Central CDK/cyclin complexes are indicated next to the cell cycle position when they are active and the Cip/Kip CDK inhibitors are shown next to CDK/cyclin complexes that they bind. The green arrow indicates that p21 and p27 are not only inhibitors but also activators of cyclin D / CDKs.
We investigate molecular mechanisms that link diverse signalling networks to the central cell cycle control machinery. At the core of this machinery is a conserved family of protein kinases called cyclin-dependent kinases (CDKs). CDKs are activated by the binding of a positive regulatory subunit, the cyclin. Sequential activation and inactivation of specific CDK complexes is required for cell cycle progression. p21 (CDK-interacting protein, Cip1), p27 (kinase-inhibitory protein, Kip1) and p57 (Kip2) constitute one of two families of CDK inhibitors (CKI) that bind to CDKs and control CDK kinase activity. Their expression, localisation and modification play a central role in regulating CDK kinase activity, especially during phase G1 and the choice between proliferation and cell cycle exit. In addition to their canonical function in CDK kinase regulation, these inhibitors can also exert CDK-independent and cell cycle-independent functions. For example, CDK inhibitor p27 can regulate cell motility and cell migration, which links this tumour suppressor protein not only to cell proliferation but also to cancer metastasis. Interestingly, some non-canonical functions of Cip/Kip proteins, such as transcriptional control, can be both CDK-dependent and CDK-independent.
Among others, we identified CDK inhibitor protein p27Kip1. Its activity, localisation or stability can be regulated by diverse mitogen signalling pathways. We investigate how these pathways control Cip/Kip family protein expression, localisation, modification, activity or function and we study their physiological roles in normal cells and cancer cells.
p27 regulates cell cycle progression over the restriction point. Abundant p27 binds and inactivates CDKs and can prevent cell proliferation. The CDK inhibitor protein becomes unstable upon cyclin/CDK2 activation, as cells traverse the restriction point and progress towards the S-phase. A positive feedback loop couples p27 ubiquitin-dependent degradation to CDK activation (Fig. 2). The molecular mechanism that can initiate this feedback loop in the presence of abundant CDK inhibitors and thus inactive CDKs has long remained an enigma.
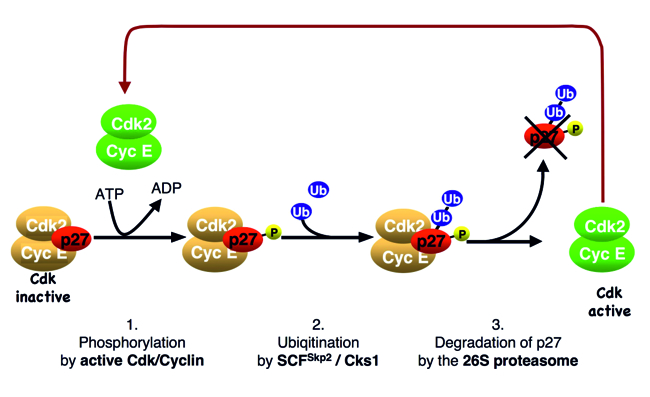
Fig. 2: A feedback loop controls CDK2 activation at the restriction point. Active CDK2 triggers the degradation of its own inhibitor, p27, which promotes the switch-like activation of CDK2 kinase at the restriction point.
We observed that oncogenic tyrosine kinases, including BCR-Abl, Src and JAK2, can activate p27-bound CDKs by phosphorylating the inhibitor. Upon phosphorylation, an inhibitory helix of p27 is ejected from the catalytic cleft of the CDK and allows the p27-bound CDK complex to bind ATP and to phosphorylate substrates. One of these substrates is p27 itself. Phosphorylation of p27 by the bound CDK generates a phosphodegron, which can initiate SCF-Skp2-mediated ubiquitin-proteasomal degradation of the CDK inhibitor (Fig. 3). Using this mechanism, which can be abused by oncogenic kinases, mitogen signals can inactivate and destabilise inhibitor p27 and thereby promote CDK activation and cell cycle progression.
Fig. 3: Oncogenic tyrosine kinases, such as JAK2, Src and BCR-Abl, can phosphorylate p27. This leads to the activation of bound CDK2. The activated CDK2 can phosphorylate bound inhibitor p27, leading to its degradation and strong CDK activation.
Ongoing research
Current research is focusing on the regulation of cell cycle progression through phase G1. We investigate mechanisms of transcriptional control by Cip/Kip proteins, translational control, temporal and spatial regulation of CDK-inhibitory proteins and the control of ubiquitin E3 ligase complexes, such as SCF-Skp2, the molecular mechanism of statin-induced cell-cycle arrest and cell-cycle control by erythropoietin (Epo) and its receptor EpoR.
Major achievements
We have discovered a novel mechanism that controls p27 stability by means of proteasomal degradation and we have identified novel pathways that control the localisation of p27. We have elucidated the molecular mechanisms that induce p27 stabilisation and Skp2 degradation in the presence of statins. We have also identified novel p27 mRNAbinding proteins, which are components of stress granules and which regulate the IRES and cap-dependent translation of p27. We have further investigated transcriptional functions of Cip/Kip protein p57Kip2, which can coactivate c-Jun/AP-1 or FHL2-mediated transcription.
Current research projects
- Function and regulation of CDK-inhibitory proteins.
- Role of translational control in the choice between cell proliferation and withdrawal from the cell cycle.
- Regulation of cytokine receptor signalling.
- Ubiquitin E3 ligase regulation in response to stress.
- Transcriptional functions of Cip/Kip proteins.
Biochemical Immunotoxicology
Johanna Gostner
Understanding of biochemical and molecular processes is a prerequisite to deciphering responses to a changing environment. Increasing exposure to environmental chemicals has an impact on human health. Nutrition and lifestyle factors may further modify susceptibilities.
Ongoing research
By integrating analytical and bio-chemistry, cell biology and immunological approaches together with biotechnological tools, we aim to investigate cellular response to chemicals, e.g. volatile compounds that affect air quality, compounds applied in everyday or care products, or phytochemicals, in order to gain a better understanding of the complex processes involved in sensitisation, irritation and adaptation. We aim to validate in vitro methods, to establish workflows for in vitro to in vivo extrapolation of effects and to evaluate biomarkers in occupational settings. Moreover, we have a keen interest in immunometabolism, primarily focusing on amino acid and pteridine biochemistry – pathways that have been shown to be susceptible to modulation, e.g. by drugs, chemicals, natural products, food or exercise. Several metabolites of the aforementioned pathways serve as sensitive and reliable biomarkers of disorders associated with chronic inflammation and provide a link between immunotoxicity, immunodeficiency and neuropsychiatric changes.
Selected Publications
- Contradictory effects of chemical filters in UV/ROS-stressed human keratinocyte and fibroblast cells. Hofer, Stefanie; Stonig, Marlies; Wally, Verena; Hartmann, Anja; Fuchs, Dietmar; Hermann, Martin; Paparella, Martin; Ganzera, Markus; Gostner, Johanna M.: ALTEX-ALTERNATIVES TO ANIMAL EXPERIMENTATION. 2019; 36(2); 231-244. PubMed: 30488083 doi: 10.14573/altex.1808201
- Homocysteine Biochemistry and Cognitive Decline in the Elderly. Gostner, Johanna; Sperner-Unterweger, Barbara; Fuchs, Dietmar: JOURNAL OF THE AMERICAN MEDICAL DIRECTORS ASSOCIATION. 2017; 18(10); 893-894. PubMed: 28847543 doi: 10.1016/j.jamda.2017.06.029
- Microbicidal activity of N-chlorotaurine can be enhanced in the presence of lung epithelial cells. Leiter, Hannes; Toepfer, Stephanie; Messner ,Petra; Rabensteiner, Marion; Gostner, Johanna, M; Lackner, Michaela; Hermann, Martin; Nagl, Markus.: JOURNAL OF CYSTIC FIBROSIS. 2020; 19(6); 1011-1017. PMID: 32201161 doi: 10.1016/j.jcf.2020.03.005. Epub 2020 Mar 20.
- Cold-inducible RNA-binding protein (CIRP) induces translation of the cell-cycle inhibitor p27Kip1. Roilo, Martina, Kullmann, Michael K., Hengst, Ludger. NUCLEIC ACIDS RESEARCH (2018). 46(6): 3192-3210.
- p27Kip1 – p(RHOB)lematic in lung cancer. Podmirseg, Silvio R.; Vosper, Jonathan; Hengst, Ludger. JOURNAL OF PATHOLOGY 2019; 3-5. doi:10.1002/path.5218
- The CDK inhibitor p57Kip2 enhances the activity of the transcriptional coactivator FHL2. Kullmann, Michael K.; Podmirseg, Silvio R.; Roilo, Martina; Hengst, Ludger, SCIENTIFIC REPORTS: 2020; April 28; 10 (1): 7140. doi:1038/s41598-020-62641-4.
- Stimulation of c-Jun/AP-1 activity by the cell cycle inhibitor p57Kip2. Kullmann, Michael K.; Pegka, Fragka; Ploner, Christian; Hengst, Ludger. Frontiers in Cell and Developmental Biology: 2021; March doi:10.3389/fcell.2021.664609.
Selection of Funding
- VocOnCell 2.0 – FFG Bridge
- Extremophiles, an innovative source for bioactive compounds - FWF (coapplicant)
- FER: Formaldehyde exposure and cellular regeneration. Cooperation with Egger
- FWF Doc-82/10 :Cellular Basis of Diseases: Molecular Control of Metabolism and Inflammation
- Coupling capillary electrophoresis to mass spectrometry for protein and proteome analysis. Industrial Project with AB SCIEX; U.S.A.
- Investigation of in-vivo O-Glycosylation of the low abundance marker NT-proBNP by Affinity Proteomics Methods and Mass Spectrometry in blood plasma from patients with severe heart failure. Industrial Project with Roche Diagnostics GmbH Penzberg, Germany
Collaborations
- Joyce M Slingerland, University of Miami, USA
- Richard W. Kriwacki, St. Jude Hosp. of Sick Children, Memphis, USA
- Frauke Melchior, Universität Heidelberg, Germany
- Pierre Roger, Universite Libre de Bruxelles, Bruxelles, Belgium
- Stephen J. Elledge, Harvard University, Boston, USA
- Joan Conaway, Stowers Institute Kansas City, USA
- Drorit Neumann, Tel Aviv University, Israel
- Terrance Lappin, Queens University Belfast, UK
Devices & Services
Protein Core Facility
The Protein Core Facility located at the Institute of Medical Biochemistry is dedicated to provide investigators with equipment, expertise and custom services for the detection, characterization and quantification of proteins and peptides on a recharge basis. The facility maintains a suite of state of the art instrumentation including different mass spectrometers (e.g. QExactive HF and LTQ Orbitrap XL from Thermo Scientific) coupled to nano-LC gradient systems and capillary electrophoresis. Also trace element analysis is provided using a Solaar M6 Dual Zeeman spectrometer (Thermo Scientific). Services include comprehensive protein identification of simple and complex protein digests, quantitative proteomics using isotope labeling strategies (e.g., SILAC, iTRAQ, TMT, Dimethyl labeling), localization and quantification of post-translational modifications (phosphorylation, acetylation, methylation, etc.), co-immunoprecipitation and affinity proteomics methods.
Head: PD Dr. Bettina Sarg
https://www.i-med.ac.at/protein-core-facility/index.html
Email: protein.facility@i-med.ac.at
 Univ.-Prof. Dr. Ludger Hengst
Univ.-Prof. Dr. Ludger Hengst
Director
Contact:
Innrain 80-82
6020 Innsbruck
Austria
Email: ludger.hengst@i-med.ac.at
Phone: +43 512 9003 70110
Fax: +43 512 9003 73100
https://www.i-med.ac.at/imcbc/medclinchemfolder/medclinchem.html



